Evaluation of Heterologous Biosynthetic Pathways for Methanol-Based 5-Aminovalerate Production by Thermophilic Bacillus methanolicus
- 1Department of Biotechnology and Food Science, Norwegian University of Science and Technology, Trondheim, Norway
- 2Department of Biotechnology and Nanomedicine, SINTEF Industry, Trondheim, Norway
- 3Toulouse Biotechnology Institute, Université de Toulouse, CNRS, INRA, INSA, Toulouse, France
The use of methanol as carbon source for biotechnological processes has recently attracted great interest due to its relatively low price, high abundance, high purity, and the fact that it is a non-food raw material. In this study, methanol-based production of 5-aminovalerate (5AVA) was established using recombinant Bacillus methanolicus strains. 5AVA is a building block of polyamides and a candidate to become the C5 platform chemical for the production of, among others, δ-valerolactam, 5-hydroxy-valerate, glutarate, and 1,5-pentanediol. In this study, we test five different 5AVA biosynthesis pathways, whereof two directly convert L-lysine to 5AVA and three use cadaverine as an intermediate. The conversion of L-lysine to 5AVA employs lysine 2-monooxygenase (DavB) and 5-aminovaleramidase (DavA), encoded by the well-known Pseudomonas putida cluster davBA, among others, or lysine α-oxidase (RaiP) in the presence of hydrogen peroxide. Cadaverine is converted either to γ-glutamine-cadaverine by glutamine synthetase (SpuI) or to 5-aminopentanal through activity of putrescine oxidase (Puo) or putrescine transaminase (PatA). Our efforts resulted in proof-of-concept 5AVA production from methanol at 50°C, enabled by two pathways out of the five tested with the highest titer of 0.02 g l–1. To our knowledge, this is the first report of 5AVA production from methanol in methylotrophic bacteria, and the recombinant strains and knowledge generated should represent a valuable basis for further improved 5AVA production from methanol.
Introduction
The worldwide amino acid market is progressively growing at 5.6% annual rate and is estimated to reach US$25.6 billion by 2022, with amino acids used for animal feed production being its largest component (Wendisch, 2020). The growing demand for amino acid supply confronts the biotechnological industry with an unprecedented challenge of identifying suitable feedstocks, especially in terms of replacing sugars and agricultural products, use whereof deteriorates food supply and threatens biodiversity (Cotton et al., 2020). Methanol, together with other one-carbon (C1) compounds, is considered a very promising substitute for feedstock that are conventionally used in biotechnological processes. The major advantages of using methanol as carbon source are its low production cost (e.g., methanol from steam reforming of methane), ease of transport and storage, and complete miscibility that bypasses the mass transfer barrier and potentially supports improvement in microbial productivities. However, what seems to cause a considerable difficulty in propagation of methanol as biotechnological feedstock is the limited selection of microorganisms capable to be used as their carbon and energy source. One of the compelling candidates to become a workhorse for the methanol-based production of amino acids is Bacillus methanolicus, a thermophilic methylotroph isolated from freshwater marsh soil by Schendel et al. (1990). The wild-type strain MGA3 naturally overproduces L-glutamate in methanol-controlled fed-batch fermentations with volumetric titers reaching up to 60 g l–1 (Heggeset et al., 2012; Table 1). Furthermore, thanks to recent developments in the toolbox for gene overexpression, it was engineered for production of different amino acid derivatives such as γ-aminobutyric acid and cadaverine (Nærdal et al., 2015; Irla et al., 2017; Table 1). MGA3 produces 0.4 g l–1 of L-lysine in high cell density fed-batch fermentations (Brautaset et al., 2010; Table 1); this titer was improved nearly 30-fold up to 11 g l–1 by plasmid-based overexpression of a gene coding for aspartokinase, a key enzyme controlling the synthesis of aspartate-derived amino acids (Jakobsen et al., 2009). Through application of a classical mutagenesis technique, a derivative of B. methanolicus MGA3 (M168-20) was constructed, which produces 11 g l–1 of L-lysine in high cell density methanol-controlled fed-batch fermentations (Brautaset et al., 2010); the L-lysine overproduction being caused among others by mutation in the hom-1 gene coding for homoserine dehydrogenase (Hom) and in the putative lysine 2,3-aminomutase gene (locus tag BMMGA3_02505). The mutation in hom-1 leads to the loss of catalytic activity of homoserine dehydrogenase and redirection of metabolic flux toward the L-lysine pathway and therefore its accumulation (Nærdal et al., 2011, 2017).

Table 1. Comparison of the 5AVA production by different engineered microbial strains and production of amino acids by B. methanolicus.
5-Aminovalerate (5AVA) is a product of L-lysine degradation, and it is mainly synthesized in a two-step process catalyzed by a lysine monooxygenase (DavB) and a δ-aminovaleramide amidohydrolase (DavA) (Revelles et al., 2005). 5AVA is a non-proteogenic five-carbon amino acid that could potentially be used as building block for producing biobased polyamides (Adkins et al., 2013; Park et al., 2014; Wendisch et al., 2018). It is also a promising precursor for plasticizers and chemicals that are intermediates for bioplastic preparation: δ-valerolactam (Chae et al., 2017), 5-hydroxy-valerate (Sohn et al., 2021), glutarate (Adkins et al., 2013; Pérez-García et al., 2018), and 1,5-pentanediol (Cen et al., 2021). As summarized in Table 1, diverse approaches have been made at the establishment of microbial 5AVA production. Pseudomonas putida KT2440, which possesses davBA in its genome, can synthesize 20.8 g l–1 5AVA from 30 g l–1 L-lysine in 12 h (Liu et al., 2014). Production of 5AVA was established in Corynebacterium glutamicum by heterologous overexpression of the DavB- and DavA-encoding genes (davBA) from P. putida with a final titer up to 39.9 g l–1 in a sugar-based fed-batch fermentation (Rohles et al., 2016; Shin et al., 2016; Joo et al., 2017). 5AVA can be also produced in a process of bioconversion of L-lysine supplemented to the growth medium with molar yields of up to 0.942 achieved by Escherichia coli strains overproducing DavBA (Park et al., 2014; Wang et al., 2016). Moreover, when the recombinant E. coli strain expressing davAB genes was cultured in a medium containing 20 g l–1 glucose and 10 g l–1 L-lysine, 3.6 g l–1 5AVA was produced, representing a molar yield of 0.45 (Park et al., 2013). Disruption of native lysine decarboxylase (CadA and LdcC) activity in E. coli strains overexpressing davBA limited cadaverine by-product formation, enabling increased accumulation of L-lysine following 5AVA production, with 5AVA yield of 0.86 g l–1 in glucose-based shaking flask fermentation (Adkins et al., 2013). Furthermore, Cheng et al. (2018) reported that the oxidative decarboxylation of L-lysine catalyzed by a L-lysine α-oxidase (RaiP) from Scomber japonicus led to 5AVA production. The production of RaiP was enhanced by the addition of 4% (v/v) ethanol and 10 mM H2O2, which increased the 5AVA titer to 29.12 g l–1 by an E. coli host strain in a fed-batch fermentation (Cheng et al., 2018). Recently, in a similar L-lysine bioconversion strategy, an E. coli whole-cell catalyst producing RaiP was developed, converting 100 g l–1 of L-lysine hydrochloride to 50.62 g l–1 5AVA representing a molar yield of 0.84 (Cheng et al., 2020).
Recent efforts have employed novel metabolic routes toward 5AVA. In Pseudomonas aeruginosa PAO1, the set of enzymes composed of glutamylpolyamine synthetase, polyamine:pyruvate transaminase, aldehyde dehydrogenase, and glutamine amidotransferase is essential for the degradation of diamines through the γ-glutamylation pathway (Yao et al., 2011), which may lead to 5AVA production when cadaverine is degraded (Luengo and Olivera, 2020). Jorge et al. (2017) established a three-step 5AVA biosynthesis pathway consisting of the conversion of L-lysine to cadaverine by the activity of the enzyme LdcC, followed by cadaverine conversion to 5AVA through consecutive transamination, by a putrescine transaminase (PatA), and oxidation by a PatD. The heterologous overexpression of the genes ldcC, patA, and patD led to 5AVA production to a final titer of 5.1 g l–1 by an engineered C. glutamicum strain in a shake flask fermentation (Jorge et al., 2017). This pathway has served as basis for the establishment of a new three-step pathway toward 5AVA using the monooxygenase putrescine oxidase (Puo), which catalyzes the oxidative deamination of cadaverine, instead of PatA (Haupka et al., 2020).
Critical factors that can affect 5AVA accumulation in a production host are the presence of a native 5AVA degradation pathway in its genome and the end product-related inhibition. In some bacterial species, such as P. putida KT2440, Pseudomonas syringae, Pseudomonas stutzeri, and C. glutamicum, 5AVA is degraded by a GABAse (Figure 1), composed of two enzymes γ-aminobutyric acid aminotransferase (GabT) and succinic semialdehyde dehydrogenase (GabD) (Park et al., 2013; Rohles et al., 2016; Pérez-García et al., 2018); for example, GABAse from Pseudomonas fluorescens KCCM 12537 retains 47.7% activity when 5AVA is used as its substrate in comparison to when GABA is used (So et al., 2013). Based on the previous research, B. methanolicus seems a feasible candidate for 5AVA production because it does not possess the necessary genetic background for GABAse-based 5AVA degradation, lacking the gabT gene in its genome (Irla et al., 2017). It was reported that 5AVA does not supports growth of B. methanolicus neither as sole carbon source nor as sole nitrogen source (Haupka et al., 2021). However, B. methanolicus displays low tolerance to 5AVA, with growth being impaired by addition of 1.17 g l–1 5AVA to the culture broth (Haupka et al., 2021).
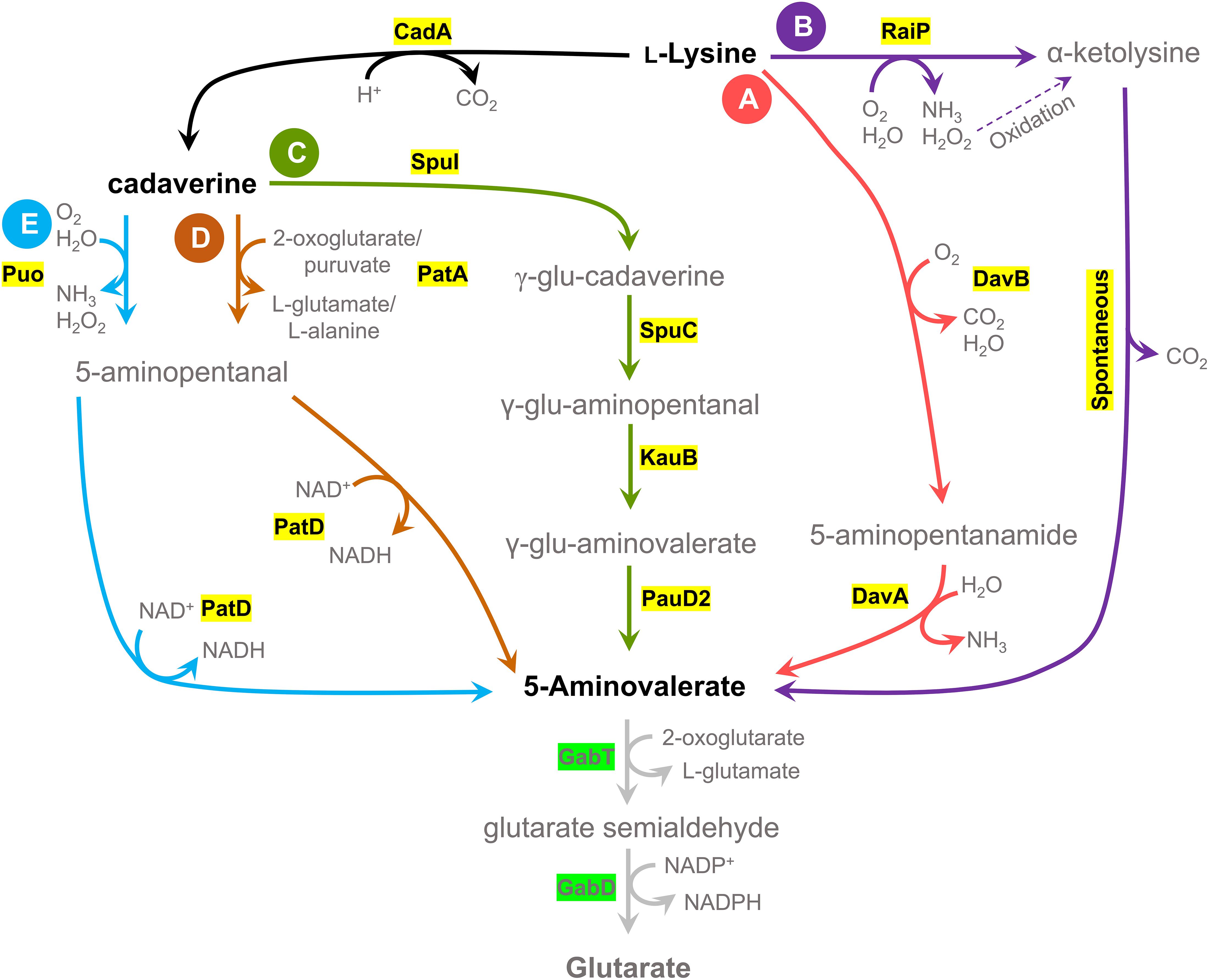
Figure 1. Schematic view of five 5AVA biosynthesis pathways and a 5AVA degradation pathway. Five different pathways for potential 5AVA production in Bacillus methanolicus were tested; two pathways have L-lysine as precursor, and three pathways have cadaverine as an intermediate metabolite, obtained by conversion of L-lysine by a lysine decarboxylase (CadA). (A) DavBA pathway: L-lysine conversion to 5AVA by lysine 2-monooxygenase (DavB) and 5-aminovaleramidase (DavA). (B) RaiP pathway: conversion of L-lysine to α-ketolysine by a L-lysine α-oxidase (RaiP) and spontaneous decarboxylation of α-ketolysine in the presence of hydrogen peroxide. (C) SpuI pathway: cadaverine to γ-glutamine-cadaverine (γ-glu-cadaverine) by glutamylpolyamine synthetase (SpuI), with subsequent activity of polyamine:pyruvate transaminase (SpuC), aldehyde dehydrogenase (KauB), and glutamine amidotransferase class I (PauD2); γ-glu-aminopentanal: γ-glutamine-aminopentanal, γ-glu-aminovalerate: γ-glutamine-aminovalerate. (D) PatA pathway: cadaverine to 5-aminopentanal through activity of putrescine aminase (PatA) and 5-aminopentanal conversion to 5AVA by 5-aminopentanal dehydrogenase (PatD). (E) Puo pathway: cadaverine to 5-aminopentanal through activity of putrescine oxidase (Puo), followed by 5AVA formation by PatD. 5AVA is degraded to glutarate by GABAse activity, a combination of γ-aminobutyrate aminotransferase (GabT) and succinate semialdehyde dehydrogenase (GabD), although this activity was not found in B. methanolicus (Irla et al., 2017).
Even though the application of diverse 5AVA biosynthetic pathways has led to significant improvement in titers and yields of 5AVA production in bacterial hosts, the most efficient processes rely on raw materials that contain sugar and/or agricultural products. Addressing shortages of global resources and food requires a replacement of the current mode of industrial biotechnology, which results in the need for novel biosynthetic pathways that utilize alternative raw materials such as methanol. Hence, in the present study we have selected five different pathways to establish methanol-based 5AVA production in the methylotrophic bacterium B. methanolicus. For two of the five pathways, proof-of-principle 5AVA production was achieved and our results should represent a valuable basis of knowledge and strains for further improved 5AVA production from methanol at 50°C.
Materials and Methods
Retrosynthesis Analysis
Retrosynthesis analysis was conducted with RetroPath 2 (Delépine et al., 2018) (v6) and RetroRules (Duigou et al., 2019) (1.0.2, with hydrogens, in reversed direction) that translated reactions from MetaNetX (Moretti et al., 2016) into reaction rules, and KNIME (3.6.1). The “source” used in this analysis was 5AVA [InChI = 1S/C5H11NO2/c6-4-2-1-3-5(7)8/h1-4,6H2,(H,7,8)], and “sink” was the set of all metabolites from E. coli genome-scale model iJO1366 (Orth et al., 2011). We used at most four reaction steps and a diameter of eight chemical bonds around the reaction center. Those conservative parameters were used to limit the strength of the substrate promiscuity hypothesis and to limit our results to pathways most likely to compete with known pathways.
Strains, Genomic DNA, Plasmids, and Primers
Bacterial strains and plasmids used in this study are listed in Table 2. The E. coli strain DH5α was used as general cloning host, and B. methanolicus strains MGA3 and M168-20 were used as expression hosts. The following strains were the source of genetic material for cloning of the 5AVA synthesis pathways: E. coli MG1655, Rhodococcus qingshengii DSM45257, Paenarthrobacter aurescens DSM20116, Kocuria rosea DSM20447, Peribacillus simplex DSM1321, and P. putida KT2440. The L-lysine-α-oxidase-coding regions from Trichoderma viride (GenBank AB937978.1) and S. japonicus (GenBank AB970726.1) were codon-optimized for B. methanolicus MGA3 expression and synthesized by Twist Biosciences (Supplementary Table S1 and Supplementary Material). The davBA operons from alternative hosts Williamsia sterculiae CPCC 203464, Roseobacter denitrificans OCh 114 strain DSM 7001, and Parageobacillus caldoxylosilyticus B4119 (davA only) were codon-optimized for expression in B. methanolicus, synthesized and provided in the pUC57 plasmid from GenScript (Supplementary Table S1 and Supplementary Material). Isolated genomic DNA of Bacillus megaterium DSM32 was purchased from German Collection of Microorganisms and Cell Cultures GmbH (DSMZ). All primers (Sigma-Aldrich) used in this research are listed in Table 2.
Molecular Cloning
The E. coli DH5α competent cells were prepared according to the calcium chloride protocol as described in Green and Rogers (2013) or purchased as chemically competent NEB 5-α E. coli cells (New England Biolabs). All standard molecular cloning procedures were carried out as described in Sambrook and Russell (2001) or according to manuals provided by producers. Chromosomal DNA was isolated as described in Eikmanns et al. (1994). PCR products were amplified using CloneAmp HiFi PCR Premix (Takara) and purified using a QIAquick PCR Purification Kit from Qiagen. DNA fragments were separated using 8 g l–1 SeaKem LE Agarose gels (Lonza) and isolated using a QIAquick Gel Extraction Kit (Qiagen). The colony PCR was performed using GoTaq DNA Polymerase (Promega). The sequences of cloned DNA fragments were confirmed by Sanger sequencing (Eurofins). B. methanolicus MGA3 was made electrocompetent and transformed by electroporation as described previously (Jakobsen et al., 2006). Recombinant DNA was assembled in vitro by means of the isothermal DNA assembly method (Gibson et al., 2009), employing the NEBuilder HiFi DNA Assembly Kit or ligation with T4 DNA ligase. pMI2mp plasmid was obtained via site-directed mutagenesis (SDM) of pTH1mp performed as previously described with CloneAmp HiFi PCR Premix (Liu and Naismith, 2008). The detailed description of plasmid creation is presented in Supplementary Material.
Media and Conditions for Shake Flask Cultivations
E. coli and P. putida strains were cultivated at 37°C in Lysogeny Broth (LB) or on LB agar plates supplemented with antibiotics when necessary. P. aurescens DSM2011 and K. rosea DSM20447 were cultivated at 30°C and 225 rpm in medium 53 (casein peptone, tryptic digest, 10.0 g l–1, yeast extract, 5.0 g l–1, glucose, 5.0 g l–1, NaCl, 5.0 g l–1; pH adjusted to 7.2–7.4); R. qingshengii DSM45257 was grown at 28°C and 225 rpm in medium 65 (glucose, 4.0 g l–1, yeast extract, 4.0 g l–1, malt extract, 10.0 g l–1; adjusted to pH to 7.2); and P. simplex DSM1321 was cultivated in nutrient medium (peptone 5 g l–1 and meat extract 3 g l–1; pH adjusted to 7.0) at 30°C and 200 rpm. For preparation of crude extracts, electrocompetent cells and transformation B. methanolicus strains were cultured at 50°C in SOB medium (Difco) supplemented with antibiotics when necessary. For 5AVA production experiments, recombinant B. methanolicus strains were cultivated in 250-ml baffled shake flasks at 50°C and 200 rpm in 40 or 50 ml MVcM medium containing 200 mM methanol. The MVcM medium contained the following, in 1 l of distilled water: K2HPO4, 4.09 g; NaH2PO4∗H2O, 1.49 g; (NH4)2SO4, 2.11 g; it was adjusted to pH 7.2 before autoclaving. The MVcM medium was supplemented with 1 ml 1 M MgSO4∗7H2O solution, 1 ml trace element solution, and 1 ml vitamin solution (Schendel et al., 1990). One mole of MgSO4∗7H2O solution contained 246.47 g of MgSO4∗7H2O in 1 l of distilled water. The trace element solution contained the following, in 1 l of distilled water: FeSO4∗7H20, 5.56 g; CuSO4∗2H2O, 27.28 mg; CaCl2∗2H2O, 7.35 g; CoCl2∗6H2O, 40.50 mg; MnCl2∗4H2O, 9.90 g; ZnSO4∗7H2O, 287.54 mg; Na2MoO4∗2H2O, 48.40 mg; H3BO3, 30.92 mg; and HCl, 80 ml. The vitamin solution contained the following, in 1 l of distilled water: biotin, thiamine hydrochloride, riboflavin, D-calcium pantothenate, pyridoxine hydrochloride, nicotinamide, 0.1 g each; p-aminobenzoic acid, 0.02 g; folic acid, vitamin B12 and lipoic acid, 0.01 g each (Schendel et al., 1990). When needed, 10 g l–1 xylose (v/v) was added for induction. For precultures, a minimal medium supplemented with 0.25 g l–1 yeast extract, designated MVcMY, was used. Antibiotics (chloramphenicol, 5 μg ml–1 and/or kanamycin, 25 μg ml–1) were supplemented as necessary. Cultivations were performed in triplicates with start OD600 of 0.1–0.2. Growth was monitored by measuring OD600 with a cell density meter (WPA CO 8000 Biowave).
Determination of Amino Acid Concentration
For the analysis of amino acid concentrations, 1 ml of the culture sample was taken from the bacterial cultures and centrifuged for 10 min at 11,000 rpm. Extracellular amino acids were quantified by means of high-pressure liquid chromatography (HPLC, Waters Alliance e2695 Separations Module). The samples underwent FMOC-Cl (fluorenylmethyloxycarbonyl chloride) derivatization before the analysis, according to the protocol described before (Haas et al., 2014), and were separated on a column (Symmetry C18 Column, 100 Å, 3.5 μm, 4.6 mm × 75 mm, Waters) according to the gradient flow presented in Table 3, where A is an elution buffer 50 mM Na-acetate pH = 4.2 and B is an organic solvent, acetonitrile. The detection was performed with a Waters 2475 HPLC Multi Fluorescence Detector (Waters), with excitation at 265 nm and emission at 315 nm.
Table 3. Determined parameters of mobile phase gradient conditions in a HPLC separation of FMOC-derivatized amino acids.
Enzyme Assays
In order to determine enzymatic activity, crude extracts of recombinant B. methanolicus cells were prepared according to Drejer et al. (2020). B. methanolicus strains were inoculated in SOB medium and grown to exponential phase (OD600 = 0.8). Recombinant expression was induced by addition of 10 g l–1 xylose 2 h after inoculation. A total amount of 50 ml culture broth was harvested by centrifugation at 7,500 rpm and 4°C for 15 min and washed twice in ice-cold buffer used for specific enzyme assay before storing at −80°C. The cells were thawed in ice and disrupted by sonication using a Fisherbrand Sonic Dismembrator (FB-505) with 40% amplitude with 2 s on and 1 s off-pulse cycles for 7 min. Cell debris was then removed by centrifugation (at 14, 000 rpm and 4°C for 1 h). Protein concentrations were determined by Bradford assay (Bradford, 1976), using bovine albumin serum (Sigma) as standard.
L-Lysine α-oxidase activity was assayed by measuring the rate of hydrogen peroxide formation, as described elsewhere (Tani et al., 2015a). The reaction was initiated by adding crude extracts from B. methanolicus strains to the reaction media (50°C) consisting of 100 mM L-lysine and 50 mM pH 7 phosphate buffer, resulting in a total volume of 1 ml. Next, the sample was quenched by addition of 50 μl 2 M HCl. After neutralization with 50 μl 2 M NaOH, 200 μl of the mixture was withdrawn and transferred to 800 μl of a second reaction mixture containing 50 mM pH 6 phosphate buffer, 30 mM phenol, 2 units ml–1 peroxidase from horseradish (Sigma) and 0.5 mM 4-aminoantipyrine. Formation of quinoneimine dye from oxidative coupling of phenol and 4-aminoantipyrine (Job et al., 2002) was determined by measuring absorbance at 505 nm using a Cary 100 Bio UV-visible spectrophotometer (Varian). One unit (U) of RaiP activity was defined as the amount of enzyme that catalyzes the formation of 1 μmol hydrogen peroxide per minute.
Catalytic activities of PatA and PatD or putrescine oxidase and PatD were measured by using a coupled reaction, and cadaverine was used as substrate instead of putrescine, as previously described elsewhere, with modifications (Jorge et al., 2017). The 1-ml assay mix contained 0.1 M Tris–HCl pH 8.0, 1.5 mM α-ketoglutarate, 2.5 mM cadaverine, 0.1 mM pyridoxal-5′-phosphate, and 0.3 mM NAD. In this coupled reaction, cadaverine was converted to 5AVA via 5-aminopentanal and one unit of coupled enzyme activity was defined as the amount of the enzyme that formed 1 μmol of NADH (ε340 nm = 6.22 mM–1 cm–1) per minute at 50°C.
The coupled DavAB assay was performed as described in Liu et al. (2014) with some modifications. Five hundred microliters of crude extract was added into 50-ml Falcon tubes filled with 4 ml 100 mM phosphate buffer pH 7.0 supplemented with 10 g l–1 L-lysine. The tubes were incubated for 40 h at 30 or 50°C with stirring at 200 rpm. The samples for quantification of 5AVA concentration through HPLC (see section “Determination of Amino Acid Concentration”) were taken at the beginning of incubation, after 16 h and after 40 h.
Results and Discussion
Selection, Design, and Construction of Heterologous Biosynthetic Pathways for 5AVA Biosynthesis in B. methanolicus
Due to the fact that B. methanolicus is a thermophile, a typical issue concerning implementation of biosynthetic pathways from heterologous hosts is the lack of thermostability of the transferred enzymes. It was shown before that a screening of diverse donor organisms allows to identify pathways active at 50°C and leads to increased product titers (Irla et al., 2017; Drejer et al., 2020). In order to extend the scope of our screening, we have constructed 26 strains with five different 5AVA biosynthetic pathways, which are presented in Figure 1, derived from diverse donors. Two pathways that directly convert L-lysine to 5AVA were chosen: the DavBA pathway (Figure 1A) and the RaiP pathway (Figure 1B), as well as three pathways that use cadaverine as an intermediate: the SpuI pathway (Figure 1C), the PatA pathway (Figure 1D), and the Puo pathway (Figure 1E).
The genes encoding the core part of those pathways are cloned into a θ-replication, low copy number derivative of pHCMC04 plasmid, pBV2xp, under control of a B. megaterium-derived, xylose-inducible promoter, and the genes encoding any ancillary enzymes are cloned into pTH1mp or pMI2mp plasmids, which are compatible to pBV2xp, under control of the mdh promoter (Irla et al., 2016). The plasmids with genes encoding desired pathways were constructed as described fully in the Supplementary Material and then used to transform B. methanolicus cells leading to formation of strains presented in Table 4.
With help of retrosynthesis analysis, we have considered two pathways that utilize L-lysine directly as precursor and that utilize either DavB (EC 1.13.12.2) and DavA (EC 3.5.1.30) activity (DavBA pathway, Figure 1A) or RaiP (EC 1.4.3.14) in the presence of H2O2 (RaiP pathway, Figure 1B) for further conversion into 5AVA. For DavBA production, three different davBA operons from the following mesophilic organisms were applied: P. putida, W. sterculiae, and R. denitrificans. We could not identify a complete davBA operon from a thermophilic host; however, thermophilic P. caldoxylosilyticus possesses a putative davA gene and was also included in this study. All selected davBA operons were codon-optimized and cloned into the pBV2xp vector under control of the xylose-inducible promoter as described in the Supplementary Material. The finished vectors were used to create the following B. methanolicus strains: MGA3_DavBAPp, MGA3_DavBAWs, MGA3_DavBARd, MGA3_DavBWsAPc, and MGA3_DavAPcBRd (Table 4). Furthermore, selected davBA operons were expressed as single genes using compatible pBV2xp and pMI2mp plasmids for gene expression (Supplementary Material). The davB genes from P. putida and W. sterculiae were cloned under control of the xylose-inducible promoter in plasmid pBV2xp, while the davA gene from P. caldoxylosilyticus was cloned into the pMI2mp plasmid under control of the mdh promoter constitutively active in methylotrophic conditions. The combination of two plasmids (2p) expressing single genes resulted in creation of the following B. methanolicus strains: MGA3_DavBPpAPc(2p) and MGA3_DavBWsAPc(2p) (Table 4).
For expression of the RaiP pathway, the B. methanolicus strains MGA3_RaiPPs, MGA3_RaiPSj, and MGA3_RaiPTv (Table 4) carried heterologous raiP gene sequences from the prokaryote P. simplex and from the eukaryotic genetic donors S. japonicus and T. viride, respectively, the two latter with characterized RaiP activity (Arinbasarova et al., 2012; Tani et al., 2015a). The full length of codon-optimized sequences derived from S. japonicus and T. viride is present in the Supplementary Table S1. The original S. japonicus sequence encodes a protein with 617 amino acids and has a 52.2% GC content, while the sequence codon optimized for B. methanolicus has a GC content of 29%. The T. viride-derivative sequence was adjusted from the GC content of 42.5 to 28.6%. The substitution of nucleotides did not alter their coding amino acid sequences.
Among the pathways using cadaverine formed from L-lysine through activity of E. coli-derived lysine decarboxylase CadA (EC 4.1.1.18, encoded by cadA) as an intermediate, we considered a multistep diamine catabolic pathway of P. aeruginosa PAOI (SpuI pathway, Figure 1C) (Yao et al., 2011). In order to test this pathway for methanol-based 5AVA production, the MGA3_SpuI strain was constructed through transformation of B. methanolicus wild type with two vectors pTH1mp-cadA and pBV2xp-AVAPp, the first one carrying the cadA gene and the latter the genes encoding the SpuI pathway (Table 4 and Supplementary Material). The SpuI pathway that converts cadaverine to 5AVA is composed of the following enzymes: glutamylpolyamine synthetase (EC 6.3.1.2, SpuI), polyamine:pyruvate transaminase (EC 2.6.1.113, SpuC), aldehyde dehydrogenase (EC 1.2.1.3, KauB), and glutamine amidotransferase class I (EC 6.3.5.2, PauD2) (Yao et al., 2011).
Another pathway, also predicted by our retrosynthesis analysis, potentially leading to production of 5AVA from L-lysine is a three-step pathway composed of CadA, PatA (EC 2.6.1.82, PatA), and 5-aminopentanal dehydrogenase (EC 1.2.1.19, PatD) (PatA pathway, Figure 1D). In order to test this pathway, two strains were constructed, MGA3_PatAEc and MGA3_PatABm, through transformation of B. methanolicus with pTH1mp-cadA plasmid, and pBV2xp-AVAEc or pBV2xp-AVABm, respectively (Table 4). As described in the Supplementary Material, the lysine decarboxylase-encoding gene (cadA) was placed under control of the mdh promoter in a rolling circle vector pTH1mp. The E. coli-derived patAD operon encoding previously characterized enzymes was placed under control of the xylose-inducible promoter in pBV2xp, resulting in pBV2xp-AVAEc (Samsonova et al., 2003). The genes of the patAD operon in B. megaterium were identified based on a BLAST search of its genome and were cloned into pBV2xp, yielding pBV2xp-AVABm (Altschul et al., 1990). While the existence of prior art makes it a solid candidate, we knew that its second step catalyzed by PatA may suffer from an unfavorable thermodynamic (predicted close to 0 kJ mol–1) (Noor et al., 2012).
In our study, we have also included a pathway confirmed through retrosynthesis analysis where the step of cadaverine transamination (PatA pathway, Figure 1D) is replaced by its oxidative deamination (Puo pathway, Figure 1E) because this reaction displays a more favorable thermodynamic (predicted close to −100 kl mol–1 in cell conditions) in comparison to PatA. While a cadaverine oxidase has not been identified before, it was shown that putrescine oxidase encoded by puo retains up to 14% of its maximal activity when cadaverine is used as a substrate (Okada et al., 1979; Ishizuka et al., 1993; van Hellemond et al., 2008; Lee and Kim, 2013). We have therefore decided to express three different versions of the puo gene derived from K. rosea, P. aurescens, and R. qingshengii, together with the E. coli-derived patD gene from the pBV2xp plasmid (for details see Supplementary Material), which led to creation of the following strains: MGA3_PuoKr, MGA3_PuoPa, and MGA3_PuoRq, respectively (Table 4). In order to prevent oxidative stress caused by H2O2 formation, a native gene encoding catalase was homologously expressed from pTH1mp plasmid in all constructed strains.
Testing Recombinant B. methanolicus Strains for 5AVA Production From Methanol
The plasmids designed and built as described in the above Section were used for transformation of wild-type B. methanolicus cells and resulted in the creation of 26 different strains (Table 4) which were then tested for their ability to synthetize 5AVA. All strains were cultivated in minimal medium supplemented with methanol as the sole carbon and energy source, and the 5AVA titer was evaluated after the strains had reached the stationary growth phase as described in the following sections.
Expression of the DavAB-Encoding Genes Resulted in no 5AVA Biosynthesis in B. methanolicus
In the first attempt, we heterologously expressed genes encoding the DavBA pathway in B. methanolicus MGA3 (Figure 1A). In addition to the well-known davBA operon from P. putida (gamma-proteobacteria), the alternative davBA operon from W. sterculiae (actinobacteria) and davAB from R. denitrificans (alpha-proteobacteria) were tested for 5AVA formation in B. methanolicus MGA3. Moreover, the only enzyme identified from a thermophilic host, DavA from P. caldoxylosilyticus (bacilli), was combined with the before mentioned lysine 2-monooxygenases (DavB). P. caldoxylosilyticus has a reported optimum growth temperature from 50 to 65°C (Fortina et al., 2001).
Several considerations were made with regard to strain design, namely, adjusting the GC content and the types of codons present in the open reading frames in the genomic DNA of a donor and designing suitable expression cassettes. In total, seven different B. methanolicus strains were constructed: MGA3(pBV2xp-davBAPp) named MGA3_DavBAPp, MGA3(pBV2xp-davBAWs) named MGA3_DavBAWs, MGA3(pBV2xp-davBARd) named MGA3_DavBARd, MGA3(pBV2xp-davBWs-davAPc) named MGA3_DavBWsAPc, MGA3(pBV2xp-davAPc-davBRd) named MGA3_DavAPcBRd, MGA3(pMI2mp-davAPc)(pBV2xp-davBPp) named MGA3_DavBPpAPc(2p), MGA3(pMI2mp-davAPc) (pBV2xp-davBWs) named MGA3_DavBWsAPc(2p) (Table 4). However, in none of the tested strains (MGA3_DavBAPp, MGA3_DavBAWs, MGA3_DavBARd, MGA3_DavBWsAPc, MGA3_DavAPcBRd), the active pathway was expressed; and followingly no 5AVA accumulation was observed during shake flask cultivations in any constructed strain (data not shown). The first reaction step from L-lysine to 5-aminopentanamide requires O2 (Figure 1A), and due to the high O2 demand to facilitate the assimilation of methanol, we also tested 5AVA formation from the alternative carbon source mannitol. Neither was this strategy successful. Furthermore, the DavAB pathway was also tested in the genetic background of L-lysine-overproducing B. methanolicus strain M160-20. Specifically, the following strains were constructed: M168-20_DavBAPp, M168-20_DavAPpBPp(2p), and M168-20_DavAPpBWs(2p); however, none of them produced any detectable 5AVA (data not shown). Taken together, the DavBA pathway did not enable 5AVA formation. It is not clear whether this was caused by low enzymatic stability at 50°C (only P. caldoxylosilyticus is known to be thermophilic among the organisms found to be source organisms for the two genes). In order to exclude the effect of elevated temperature on the DavAB activity, we tested enzymatic activity at 30°C for selected strains (MGA3_DavBAPp, MGA3_DavBAWs, MGA3_DavBARd, MGA3_DavBWsAPc, and MGA3_DavAPcBRd); however, no DavAB activity was detected (data not shown). The reason why the functional DavAB pathway was not expressed in B. methanolicus remains unknown.
RaiP Pathway Is Functional in B. methanolicus and Supports 5AVA Production
Methanol-based 5AVA biosynthesis was attempted via heterologous expression of RaiP encoding gene raiP in MGA3. The strains MGA3(pBV2xp-raiPPs) named MGA3_RaiPPs, MGA3(pBV2xp-raiPSj) named MGA3_RaiPSj, and MGA3(pBV2xp-raiPTv) named MGA3_RaiPTv (Table 4) carry the raiP gene from the bacterium P. simplex and raiP genes with codon-optimized sequences from the eukaryotic donors S. japonicus and T. viride, respectively. The T. viride-derived RaiP was shown to be stable at temperatures up to 50°C (Arinbasarova et al., 2012). It is reported that the RaiP protein from S. japonicus is thermally stable for at least 1 h in temperatures up to 60°C, with its highest activity registered at 70°C (Tani et al., 2015b). Moreover, although there is no kinetic characterization of RaiP from P. simplex available, this bacterium is classified as mesophilic, with growth optimum at 30°C (Yumoto et al., 2004). To examine the activity of RaiP in the constructed B. methanolicus strains, L-lysine α-oxidase activity was measured at 50°C. While the empty vector control strain has shown no RaiP activity, the highest RaiP specific activity was observed in crude extracts from strain MGA3_RaiPTv, being 62.1 ± 1.4 mU mg–1 (Figure 2A). The values of RaiP activity for strains MGA3_RaiPPs and MGA3_RaiPSj were 1.4 ± 0.3 mU mg–1 and 12.0 ± 4.4 mU mg–1, respectively (Figure 2A). It is not clear if the poor activity of heterologous RaiP from genetic donors S. japonicus and P. simplex was caused by low enzymatic stability at 50°C, and the reason for that remains to be investigated.
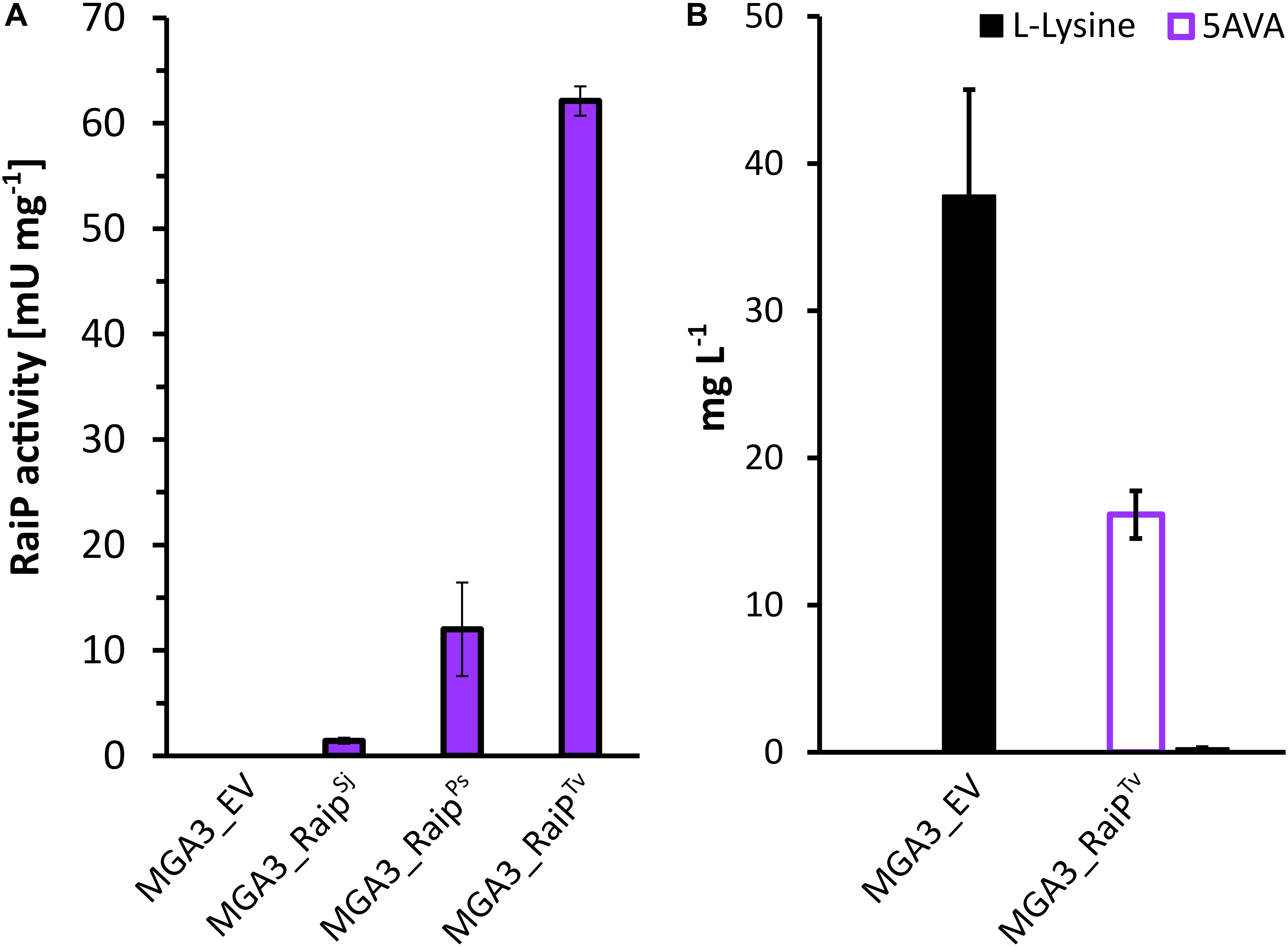
Figure 2. Evaluation of RaiP enzyme activity (A) and amino acids production (B) in recombinant B. methanolicus strains. B. methanolicus strains MGA3_EV, MGA3_RaiPSj, MGA3_RaiPPs, or MGA3_RaiPTv were cultivated in a shaking flask culture. The grown cells were harvested, washed twice with 50 mM phosphate buffer (pH 7.0), and disrupted by sonication. After centrifugation, the crude extracts were directly used for the RaiP assay. MGA3_EV and MGA3_RaiPTv were cultivated for 27 h, and supernatants were obtained by centrifugation for HPLC analysis. The error bars represent standard deviation of technical triplicates.
HPLC analysis of supernatant from MGA3_RaiPTv strain cultivated in minimal medium revealed 16.15 ± 1.62 mg L–1 5AVA and 0.27 ± 0.04 mg L–1 L-lysine. In contrast, the L-lysine level in the MGA3 strain harboring the empty vector plasmid pBV2xp (MGA3_EV) was 37.8 ± 7.2 mg L–1 (Figure 2B). Even though a slight RaiP activity was observed in crude extract of the strains MGA3_RaiPPs and MGA3_RaiPSj, no 5AVA production was observed for those strains (data not shown). Let us note here that the 5AVA titer in the methanol-based shaking flask fermentation of strain MGA3_RaiPTv was significantly inferior to that in previously reported glucose-based fermentations in E. coli (Cheng et al., 2018).
The value of the Michaelis–Menten constant for T. viride-derived RaiP for L-lysine has been estimated (Km = 5.85 mg L–1) (Kusakabe et al., 1980). Therefore, the precursor levels in the B. methanolicus strains should not be a limiting factor for production of 5AVA. The RaiP-mediated production is mainly utilized in the L-lysine bioconversion approach, utilizing E. coli strains as whole-cell biocatalysts (Cheng et al., 2018, 2020, 2021) where high concentrations of the precursor were used; for example, the molar yield of 0.942 was obtained from 120 g l–1 L-lysine (Park et al., 2014). However, construction and testing of the B. methanolicus strains M168-20_RaiPSj, M168-20_RaiPPs, and M168-20_RaiPTv (Table 4), based on the L-lysine-over producing mutant M168-20 (Brautaset et al., 2010), did not result in any improved 5AVA production (data not shown).
The lack of 5AVA production in MGA3_RaiPPs and MGA3_RaiPSj, as well as low 5AVA titer produced by strain MGA3_RaiPTv, might be related to the spontaneous conversion step that follows RaiP activity. This could be a limiting factor for the RaiP-mediated production of 5AVA. Three compounds are produced in a reaction catalyzed by RaiP: α-ketolysine, NH3, and H2O2 (Mai-Prochnow et al., 2008; Cheng et al., 2018). In a second spontaneous step of 5AVA synthesis, the intermediate α-ketolysine is oxidatively decarboxylated to form 5AVA in the presence of H2O2 as an oxidizing agent. It was shown that the addition of H2O2 into the culture broth has led to an 18−fold increase of 5AVA titers in comparison with the control condition without H2O2 (final titer 29.12 g l–1) in a 5–l fermenter (Cheng et al., 2018). The RaiP-mediated 5AVA production may be increased by enzymatic conversion of α-ketolysine in an approach different to ours, where spontaneous reaction of oxidative decarboxylation occurs. Recently, an artificial synthetic pathway for the biosynthesis of 5AVA in E. coli was developed, consisting of three steps: conversion of L-lysine to α-ketolysine via RaiP, decarboxylation of α-ketolysine to produce 5-aminopentanal via α-ketoacid decarboxylase, and oxidation of 5-aminopentanal to 5AVA via aldehyde dehydrogenase. The expression of the artificial pathway resulted in a yield increase of 774% compared to the single gene pathway (Cheng et al., 2021). This approach is potentially a feasible strategy we have shown in our study that E. coli-derived PatD is active as a 5-aminopentanal dehydrogenase in B. methanolicus and participates in 5AVA biosynthesis (see Section “The PatA Pathway Supports 5AVA Accumulation in B. methanolicus).
Use of the SpuI Pathway Does Not Lead to 5AVA Production in B. methanolicus
Three different pathways that use cadaverine as an intermediate product have been tested for their feasibility for production of 5AVA in B. methanolicus. Cadaverine biosynthesis in B. methanolicus cells was enabled through the activity of lysine decarboxylase encoded by a heterologously expressed cadA (Nærdal et al., 2015). Cadaverine can be converted to 5AVA through activity of a multistep diamine catabolic pathway derived from P. aeruginosa PAOI (SpuI pathway, Figure 1C) (Yao et al., 2011). The MGA3(pTH1mp-cadA)(pBV2xp-AVAPp) strain called MGA3_SpuI (Table 4) did not accumulate any 5AVA during methanol-based growth in minimal medium, despite the accumulation of the precursor, cadaverine, at the level of 118.8 ± 5.1 mg l–1 similar to the empty vector control strain (130.0 ± 5.3 mg l–1) (Table 5). The cadaverine titers of 130.0 ± 5.3 mg l–1 achieved by MGA3_Cad are higher than the L-lysine titer of 37.8 ± 7.2 mg L–1 achieved by MGA3_EV in this study (Figure 2B). This is in accordance with previous findings of Nærdal et al. (2011, 2015) who attributed high cadaverine titers for production strain in relation to L-lysine titer in empty vector control strain to a metabolic pull which deregulated flux through the L-lysine biosynthesis pathway.
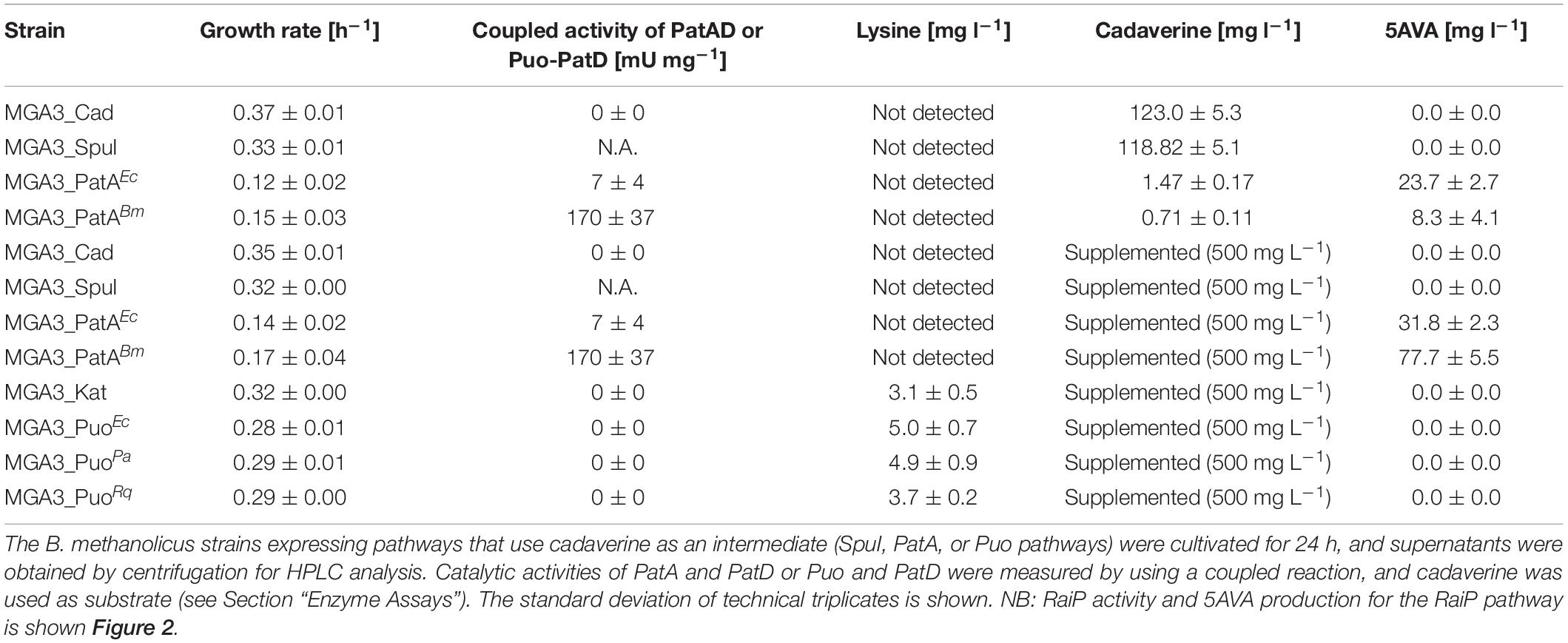
Table 5. Growth rates, enzyme activities and L-lysine, cadaverine, and 5AVA final titers accumulated in growth media of recombinant MGA3 strains.
The PatA Pathway Supports 5AVA Accumulation in B. methanolicus
In the next step, two versions of the PatA pathway (Figure 1D) derived from either E. coli or B. megaterium were tested in strains MGA3(pTH1mp-cadA)(pBV2xp-AVAEc) named MGA3_PatAEc and MGA3(pTH1mp-cadA)(pBV2xp-AVABm) named MGA3 _PatABm (Table 4), respectively. The optimal temperature of PatA derived from E. coli is 60°C, which means that it is a thermostable enzyme that should be active at 50°C, which is a temperature used for the production experiment. PatA was shown to have a broad substrate range including cadaverine and, in lower extent, spermidine, but not ornithine (Samsonova et al., 2003). This property was used by Jorge et al. (2017) who have shown in their study that it is possible to use PatA and PatD derived from E. coli to establish conversion of cadaverine to 5AVA, confirming experimentally the broad substrate range of those two enzymes. The B. megaterium-derived PatA was characterized only superficially with regard to its substrate spectrum and not optimal temperature or thermostability (Slabu et al., 2016); however, its host organism is known to have a wide temperature range for growth up to 45°C (Vary et al., 2007). The multiple-sequence alignment with E. coli-derived enzymes showed identity of 63 and 38% for PatA and PatD, respectively (Okada et al., 1979). Both E. coli and B. megaterium-derived versions of the pathway are functional in B. methanolicus, with the combined PatAD activity of 7 ± 4 mU and 170 ± 37 mU mg–1 (Table 5). Final 5AVA titers of 23.7 ± 2.7 and 8.3 ± 4.1 mg L–1 (Table 5) were achieved, which is considerably lower than 5AVA titers of 0.9 g l–1 obtained by wild-type C. glutamicum strain transformed with plasmids for expression of ldcC (coding for lysine decarboxylase) and patDA (Jorge et al., 2017). For both producer strains, the concentration of unconverted cadaverine is similar: 1.7 ± 0.1 mg l–1 and 1.5 ± 0.2 mg l–1 for MGA3_PatAEc and MGA3_PatABm, respectively (Table 5). While Km for cadaverine has not been assessed, it has been shown to be 811 mg l–1 for putrescine for E. coli-derived PatA; assuming similar Km for cadaverine, it may explain why full conversion of cadaverine has not occurred (Samsonova et al., 2003). Due to relatively high Km for putrescine of PatA, we decided to test how supplementation with external cadaverine affects 5AVA accumulation. In fact, for both MGA3_PatAEc and MGA3_PatABm, 5AVA titers increased to 31.8 ± 2.3 and 77.7 ± 5.5, respectively, when the growth medium was supplemented with 500 mg l–1 cadaverine (Table 5). These results indicate that the enhancement of precursor supply is one potential target for subsequent metabolic engineering efforts to increase 5AVA titers. Another important consideration for activity of transaminase is availability of keto acid that acts as amino group acceptor. It was shown that E. coli and B. megaterium-derived PatA can use either pyruvate or 2-oxoglutarate as amino group acceptors (Slabu et al., 2016); the intracellular concentrations of those compounds in B. methanolicus MGA3 cells are 3.2 and 2.7 mM, respectively (Brautaset et al., 2003). Knowing that Km for 2-oxoglutarate for E. coli-derived PatA is 19.0 mM (Samsonova et al., 2003), recovery of the keto acids may be a limitation for 5AVA accumulation. This issue could be potentially solved by heterologous production of alanine dehydrogenase or L-glutamate oxidase which catalyzes reactions where pyruvate or 2-oxoglutarate is produced (Böhmer et al., 1989; Sakamoto et al., 1990; Slabu et al., 2016).
Use of the Puo Pathway Leads to 5AVA Production in B. methanolicus
Lastly, a pathway that relies on an activity of the monooxygenase putrescine oxidase (Puo, EC 1.4.3.10) was tested (Figure 1E). Puo catalyzes the oxidative deamination of cadaverine in lieu of cadaverine transamination catalyzed by PatA. It was shown that different putrescine oxidases can use cadaverine as their substrate with 9–14% of their maximal activity shown when putrescine is a substrate (Desa, 1972; Okada et al., 1979; van Hellemond et al., 2008; Lee et al., 2013). Moreover, putrescine oxidases derived from K. rosea (Micrococcus rubens) and Rhodococcus are thermostable and optimal activity of P. aurescens-derived Puo is at 50°C (Desa, 1972; van Hellemond et al., 2008; Lee et al., 2013). The disadvantage of this pathway is that it requires O2, the supply of which may be difficult to control. Furthermore, due to formation of hydrogen peroxide in the reaction catalyzed by Puo, the oxidative stress may increase when this pathway is active. In order to avoid detrimental effect of hydrogen peroxide accumulation, catalase was overproduced in the recombinant strains containing the Puo pathway: MGA3(pTH1mp-katA)(pBV2xp-AVAKr) named MGA3_PuoKr, MGA3(pTH1mp-katA)(pBV2xp AVAPa) named MGA3_PuoPa, and MGA3(pTH1mp-katA)(pBV2xp-AVARq) named MGA3_PuoRq (Table 4). To achieve sufficient levels of the pathway precursor, cadaverine, we have decided not to rely on plasmid-based production of lysine decarboxylase and to add cadaverine to the growth medium, instead. The tested recombinant strains with the Puo pathway did not produce 5AVA, which is consistent with no Puo-PatD activity detected in crude extracts (Table 5). The Puo pathway was shown to be active in C. glutamicum where titer of 0.1 ± 0.0–0.4 ± 0.0 g l–1 5AVA was achieved (Haupka et al., 2020).
Conclusion
In the search for 5AVA production from the sustainable feedstock methanol, we have screened five pathways toward 5AVA biosynthesis in B. methanolicus. No 5AVA production was observed for DavBA, Puo, and SpuI pathways. However, the pathways relying on RaiP and PatA activities were functional in shake flask cultures of B. methanolicus, which led to 5AVA production from methanol for the first time, respectively, up to 16.15 ± 1.62 mg l–1 or 23.7 ± 2.7. RaiP and PatA pathways are targets for further optimizations which could increase the 5AVA titers in the constructed strains. For instance, the improvement of substrate utilization and H2O2 availability or decomposition efficiency might contribute to the increase in the yield of 5AVA. Moreover, our study shows that the availability of supplemented cadaverine has high impact on 5AVA titer when the PatA pathway is employed. Another factor that needs to be considered is tolerance to 5AVA, which was shown to be low (Haupka et al., 2021). Recently, adaptative laboratory evolution experiments resulted in the selection of a mutant strain of B. methanolicus that displays tolerance to approximately 46 g l–1 5AVA (Haupka et al., 2021), which could be employed as a platform to develop high-titer 5AVA production strains. This shows that methanol has the potential to become a sustainable feedstock for the production of 5AVA.
Data Availability Statement
The original contributions generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LB, MI, IN, and SL: study design and experimental work. BD: bioinformatic analysis. LB and MI: writing—original draft preparation. TB: writing—review and editing and project administration. SH, TB, MI, and IN: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Research Council of Norway within ERA CoBioTech, an ERA-Net Cofund Action under H2020, grant number 285794 ERA-NET.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tonje Marita Bjerkan Heggeset from SINTEF Industry for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.686319/full#supplementary-material
References
Adkins, J., Jordan, J., and Nielsen, D. R. (2013). Engineering Escherichia coli for renewable production of the 5-carbon polyamide building-blocks 5-aminovalerate and glutarate. Biotechnol. Bioeng. 110, 1726–1734. doi: 10.1002/bit.24828
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arinbasarova, A. Y., Ashin, V. V., Makrushin, K. V., Medentsev, A. G., Lukasheva, E. V., and Berezov, T. T. (2012). Isolation and properties of L-lysine-α-oxidase from the fungus Trichoderma cf. aureoviride RIFAI VKM F-4268D. Microbiology 81, 549–554. doi: 10.1134/S0026261712050037
Böhmer, A., Müller, A., Passarge, M., Liebs, P., Honeck, H., and Müller, H.-G. (1989). A novel L-glutamate oxidase from Streptomyces endus. Eur. J. Biochem. 182, 327–332. doi: 10.1111/j.1432-1033.1989.tb14834.x
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Brautaset, T., Jakobsen, Ø. M., Degnes, K. F., Netzer, R., Nśrdal, I., Krog, A., et al. (2010). Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for L-lysine production from methanol at 50°C. Appl. Microbiol. Biotechnol. 87, 951–964. doi: 10.1007/s00253-010-2559-6
Brautaset, T., Williams, M. D., Dillingham, R. D., Kaufmann, C., Bennaars, A., Crabbe, E., et al. (2003). Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in L-lysine-secreting mutants. Appl. Microbiol. Biotechnol. 69, 3986–3995. doi: 10.1128/AEM.69.7.3986-3995.2003
Cen, X., Liu, Y., Chen, B., Liu, D., and Chen, Z. (2021). Metabolic engineering of Escherichia coli for de novo production of 1,5-pentanediol from glucose. ACS Synth. Biol. 10, 192–203. doi: 10.1021/acssynbio.0c00567
Chae, T. U., Ko, Y.-S., Hwang, K.-S., and Lee, S. Y. (2017). Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams. Metab. Eng. 41, 82–91. doi: 10.1016/j.ymben.2017.04.001
Cheng, J., Luo, Q., Duan, H., Peng, H., Zhang, Y., Hu, J., et al. (2020). Efficient whole-cell catalysis for 5-aminovalerate production from L-lysine by using engineered Escherichia coli with ethanol pretreatment. Sci. Rep. 10:990. doi: 10.1038/s41598-020-57752-x
Cheng, J., Tu, W., Luo, Z., Gou, X., Li, Q., Wang, D., et al. (2021). A high-efficiency artificial synthetic pathway for 5-aminovalerate production from biobased L-lysine in Escherichia coli. Front. Bioeng. Biotechnol. 9:633028. doi: 10.3389/fbioe.2021.633028
Cheng, J., Zhang, Y., Huang, M., Chen, P., Zhou, X., Wang, D., et al. (2018). Enhanced 5-aminovalerate production in Escherichia coli from L-lysine with ethanol and hydrogen peroxide addition. J. Chem. Technol. Biotechnol. 93, 3492–3501. doi: 10.1002/jctb.5708
Cotton, C. A., Claassens, N. J., Benito-Vaquerizo, S., and Bar-Even, A. (2020). Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 62, 168–180. doi: 10.1016/j.copbio.2019.10.002
Delépine, B., Duigou, T., Carbonell, P., and Faulon, J. L. (2018). RetroPath2.0: a retrosynthesis workflow for metabolic engineers. Metab. Eng. 45, 158–170. doi: 10.1016/j.ymben.2017.12.002
Desa, R. J. (1972). Putrescine oxidase from Micrococcus rubens. J. Biol. Chem. 247, 5527–5534. doi: 10.1016/s0021-9258(20)81137-5
Drejer, E. B., Chan, D. T. C., Haupka, C., Wendisch, V. F., Brautaset, T., and Irla, M. (2020). Methanol-based acetoin production by genetically engineered Bacillus methanolicus. Green Chem. 22, 788–802. doi: 10.1039/c9gc03950c
Duigou, T., Du Lac, M., Carbonell, P., and Faulon, J. L. (2019). Retrorules: a database of reaction rules for engineering biology. Nucleic Acids Res. 47, D1229–D1235. doi: 10.1093/nar/gky940
Eikmanns, B. J., Thum-schmitz, N., Eggeling, L., Ludtke, K., and Sahm, H. (1994). Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140, 1817–1828. doi: 10.1099/13500872-140-8-1817
Fortina, M. G., Mora, D., Schumann, P., Parini, C., Manachini, P. L., and Stackebrandt, E. (2001). Reclassification of Saccharococcus caldoxylosilyticus as Geobacillus caldoxylosilyticus (Ahmad et al. 2000) comb. nov. Int. J. Syst. Evol. Microbiol. 51(Pt 6), 2063–2071. doi: 10.1099/00207713-51-6-2063
Gibson, D., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Green, R., and Rogers, E. J. (2013). Transformation of chemically competent E. coli. Methods Enzymol. 529, 329–336. doi: 10.1016/B978-0-12-418687-3.00028-8
Haas, T., Poetter, M., Pfeffer, J. C., Kroutil, W., Skerra, A., Lerchner, A., et al. (2014). Oxidation and Amination of Secondary Alcohols. Patent number EP2734631B1. Munich: European Patent Office.
Haupka, C., Delépine, B., Irla, M., Heux, S., and Wendisch, V. F. (2020). Flux enforcement for fermentative production of 5-aminovalerate and glutarate by Corynebacterium glutamicum. Catalysts 10:1065. doi: 10.3390/catal10091065
Haupka, C., Fernandes de Brito, L., Busche, T., Wibberg, D., and Wendisch, V. F. (2021). Genomic and transcriptomic investigation of the physiological response of the methylotroph Bacillus methanolicus to 5-aminovalerate. Front. Microbiol. 12:664598. doi: 10.3389/fmicb.2021.664598
Heggeset, T. M. B., Krog, A., Balzer, S., Wentzel, A., Ellingsen, T. E., and Brautaset, T. (2012). Genome sequence of thermotolerant Bacillus methanolicus: features and regulation related to methylotrophy and production of L-lysine and L-glutamate from methanol. Appl. Environ. Microbiol. 78, 5170–5181. doi: 10.1128/AEM.00703-12
Irla, M., Heggeset, T. M. B., Nærdal, I., Paul, L., Haugen, T., Le, S. B., et al. (2016). Genome-based genetic tool development for Bacillus methanolicus: theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production. Front. Microbiol. 7:1481. doi: 10.3389/fmicb.2016.01481
Irla, M., Nærdal, I., Brautaset, T., and Wendisch, V. F. (2017). Methanol-based γ-aminobutyric acid (GABA) production by genetically engineered Bacillus methanolicus strains. Ind. Crops Prod. 106, 12–20. doi: 10.1016/j.indcrop.2016.11.050
Ishizuka, H., Horinouchi, S., and Beppu, T. (1993). Putrescine oxidase of Micrococcus rubens: primary structure and Escherichia coli. J. Gen. Microbiol. 139, 425–432. doi: 10.1099/00221287-139-3-425
Jakobsen, Ø. M., Benichou, A., Flickinger, M. C., Valla, S., Ellingsen, T. E., and Brautaset, T. (2006). Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J. Bacteriol. 188, 3063–3072. doi: 10.1128/JB.188.8.3063
Jakobsen, Ø. M., Brautaset, T., Degnes, K. F., Heggeset, T. M. B., Balzer, S., Flickinger, M. C., et al. (2009). Overexpression of wild-type aspartokinase increases L-lysine production in the thermotolerant methylotrophic bacterium Bacillus methanolicus. Appl. Environ. Microbiol. 75, 652–661. doi: 10.1128/AEM.01176-08
Job, V., Marcone, G. L., Pilone, M. S., and Pollegioni, L. (2002). Glycine oxidase from Bacillus subtilis: characterization of a new flavoprotein. J. Biol. Chem. 277, 6985–6993. doi: 10.1074/jbc.M111095200
Joo, J. C., Oh, Y. H., Yu, J. H., Hyun, S. M., Khang, T. U., Kang, K. H., et al. (2017). Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a Miscanthus hydrolysate solution prepared by a newly developed Miscanthus hydrolysis process. Bioresour. Technol. 245, 1692–1700. doi: 10.1016/j.biortech.2017.05.131
Jorge, J. M. P., Pérez-García, F., and Wendisch, V. F. (2017). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245, 1701–1709. doi: 10.1016/j.biortech.2017.04.108
Kusakabe, H., Kodama, K., Kuninaka, A., Yoshino, H., Misono, H., and Soda, K. (1980). A new antitumor enzyme, L-lysine alpha-oxidase from Trichoderma viride. Purification and enzymological properties. J. Biol. Chem. 255, 976–981. doi: 10.1016/S0021-9258(19)86128-8
Lee, J.-I., Jang, J.-H., Yu, M.-J., and Kim, Y.-W. (2013). Construction of a bifunctional enzyme fusion for the combined determination of biogenic amines in foods. J. Agric. Food Chem. 61, 9118–9124. doi: 10.1021/jf403044m
Lee, J. I., and Kim, Y. W. (2013). Characterization of amine oxidases from Arthrobacter aurescens and application for determination of biogenic amines. World J. Microbiol. Biotechnol. 29, 673–682. doi: 10.1007/s11274-012-1223-y
Liu, H., and Naismith, J. H. (2008). An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8:91. doi: 10.1186/1472-6750-8-91
Liu, P., Zhang, H., Lv, M., Hu, M., Li, Z., Gao, C., et al. (2014). Enzymatic production of 5-aminovalerate from L-lysine using L-lysine monooxygenase and 5-aminovaleramide amidohydrolase. Sci. Rep. 4:5657. doi: 10.1038/srep05657
Luengo, J. M., and Olivera, E. R. (2020). Catabolism of biogenic amines in Pseudomonas species. Environ. Microbiol. 22, 1174–1192. doi: 10.1111/1462-2920.14912
Mai-Prochnow, A., Lucas-Elio, P., Egan, S., Thomas, T., Webb, J. S., Sanchez-Amat, A., et al. (2008). Hydrogen peroxide linked to lysine oxidase activity facilitates biofilm differentiation and dispersal in several Gram-negative bacteria. J. Bacteriol. 190, 5493–5501. doi: 10.1128/JB.00549-08
Moretti, S., Martin, O., van Du Tran, T., Bridge, A., Morgat, A., and Pagni, M. (2016). MetaNetX/MNXref - reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 44, D523–D526. doi: 10.1093/nar/gkv1117
Nærdal, I., Netzer, R., Ellingsen, T. E., and Brautaset, T. (2011). Analysis and manipulation of aspartate pathway genes for L-lysine overproduction from methanol by Bacillus methanolicus. Appl. Environ. Microbiol. 77, 6020–6026. doi: 10.1128/AEM.05093-11
Nærdal, I., Netzer, R., Irla, M., Krog, A., Heggeset, T. M. B., Wendisch, V. F., et al. (2017). L-lysine production by Bacillus methanolicus: genome-based mutational analysis and L-lysine secretion engineering. J. Biotechnol. 244, 25–33. doi: 10.1016/j.jbiotec.2017.02.001
Nærdal, I., Pfeifenschneider, J., Brautaset, T., and Wendisch, V. F. (2015). Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains. Microb. Biotechnol. 8, 342–350. doi: 10.1111/1751-7915.12257
Noor, E., Bar-Even, A., Flamholz, A., Lubling, Y., Davidi, D., and Milo, R. (2012). An integrated open framework for thermodynamics of reactions that combines accuracy and coverage. Bioinformatics 28, 2037–2044. doi: 10.1093/bioinformatics/bts317
Okada, M., Kawashima, S., and Imahori, K. (1979). Substrate specificity and reaction mechanism of putrescine oxidase. J. Biochem. 86, 97–104.
Orth, J. D., Conrad, T. M., Na, J., Lerman, J. A., Nam, H., Feist, A. M., et al. (2011). A comprehensive genome-scale reconstruction of Escherichia coli metabolism-2011. Mol. Syst. Biol. 7:535. doi: 10.1038/msb.2011.65
Park, S. J., Kim, E. Y., Noh, W., Park, H. M., Oh, Y. H., Lee, S. H., et al. (2013). Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. Metab. Eng. 16, 42–47. doi: 10.1016/j.ymben.2012.11.011
Park, S. J., Oh, Y. H., Noh, W., Kim, H. Y., Shin, J. H., Lee, E. G., et al. (2014). High-level conversion of L-lysine into 5-aminovalerate that can be used for nylon 6,5 synthesis. Biotechnol. J. 9, 1322–1328. doi: 10.1002/biot.201400156
Pérez-García, F., Jorge, J. M. P., Dreyszas, A., Risse, J. M., and Wendisch, V. F. (2018). Efficient production of the dicarboxylic acid glutarate by Corynebacterium glutamicum via a novel synthetic pathway. Front. Microbiol. 9:2589. doi: 10.3389/fmicb.2018.02589
Revelles, O., Espinosa-Urgel, M., Fuhrer, T., Sauer, U., and Ramos, J. L. (2005). Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440. J. Bacteriol. 187, 7500–7510. doi: 10.1128/JB.187.21.7500-7510.2005
Rohles, C. M., Gießelmann, G., Kohlstedt, M., Wittmann, C., and Becker, J. (2016). Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb. Cell Fact. 15:154. doi: 10.1186/s12934-016-0553-0
Sakamoto, Y., Nagata, S., Esaki, N., Tanaka, H., and Soda, K. (1990). Gene cloning, purification and characterization of thermostable alanine dehydrogenase of Bacillus stearothermophilus. J. Ferment. Bioeng. 69, 154–158. doi: 10.1016/0922-338X(90)90038-X
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Laboratory Press.
Samsonova, N. N., Smirnov, S. V., Altman, I. B., and Ptitsyn, L. R. (2003). Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol. 3:2. doi: 10.1186/1471-2180-3-2
Schendel, F. J., Bremmon, C. E., Flickinger, M. C., and Guettler, M. (1990). L-lysine production at 50°C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 56, 963–970. doi: 10.1128/AEM.56.4.963-970.1990
Shin, J. H., Park, S. H., Oh, Y. H., Choi, J. W., Lee, M. H., Cho, J. S., et al. (2016). Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid. Microb. Cell Fact. 15:174. doi: 10.1186/s12934-016-0566-8
Slabu, I., Galman, J. L., Weise, N. J., Lloyd, R. C., and Turner, N. J. (2016). Putrescine transaminases for the synthesis of saturated nitrogen heterocycles from polyamines. ChemCatChem 8, 1038–1042. doi: 10.1002/cctc.201600075
So, J. H., Lim, Y. M., Kim, S. J., Kim, H. H., and Rhee, I. K. (2013). Co-expression of gamma-aminobutyrate aminotransferase and succinic semialdehyde dehydrogenase genes for the enzymatic analysis of gamma-aminobutyric acid in Escherichia coli. J. Appl. Biol. Chem. 56, 89–93. doi: 10.3839/jabc.2013.015
Sohn, Y. J., Kang, M., Baritugo, K.-A., Son, J., Kang, K. H., Ryu, M.-H., et al. (2021). Fermentative high-level production of 5-hydroxyvaleric acid by metabolically engineered Corynebacterium glutamicum. ACS Sustain. Chem. Eng. 9, 2523–2533. doi: 10.1021/acssuschemeng.0c08118
Tani, Y., Miyake, R., Yukami, R., Dekishima, Y., China, H., Saito, S., et al. (2015a). Functional expression of L-lysine α-oxidase from Scomber japonicus in Escherichia coli for one-pot synthesis of L-pipecolic acid from DL-lysine. Appl. Microbiol. Biotechnol. 99, 5045–5054. doi: 10.1007/s00253-014-6308-0
Tani, Y., Omatsu, K., Saito, S., Miyake, R., Kawabata, H., Ueda, M., et al. (2015b). Heterologous expression of L-lysine α-oxidase from Scomber japonicus in Pichia pastoris and functional characterization of the recombinant enzyme. J. Biochem. 157, 201–210. doi: 10.1093/jb/mvu064
van Hellemond, E. W., van Dijk, M., Heuts, D. P. H. M., Janssen, D. B., and Fraaije, M. W. (2008). Discovery and characterization of a putrescine oxidase from Rhodococcus erythropolis NCIMB 11540. Appl. Microbiol. Biotechnol. 78, 455–463. doi: 10.1007/s00253-007-1310-4
Vary, P. S., Biedendieck, R., Fuerch, T., Meinhardt, F., Rohde, M., Deckwer, W.-D., et al. (2007). Bacillus megaterium-from simple soil bacterium to industrial protein production host. Appl. Microbiol. Biotechnol. 76, 957–967. doi: 10.1007/s00253-007-1089-3
Wang, X., Cai, P., Chen, K., and Ouyang, P. (2016). Efficient production of 5-aminovalerate from L-lysine by engineered Escherichia coli whole-cell biocatalysts. J. Mol. Catal. B Enzym. 134, 115–121. doi: 10.1016/j.molcatb.2016.10.008
Wendisch, V. F. (2020). Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 58, 17–34. doi: 10.1016/j.ymben.2019.03.008
Wendisch, V. F., Mindt, M., and Pérez-García, F. (2018). Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 102, 3583–3594. doi: 10.1007/s00253-018-8890-z
Yao, X., He, W., and Lu, C. D. (2011). Functional characterization of seven γ-glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and β-alanine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 193, 3923–3930. doi: 10.1128/JB.05105-11
Yumoto, I., Hirota, K., Yamaga, S., Nodasaka, Y., Kawasaki, T., Matsuyama, H., et al. (2004). Bacillus asahii sp. nov., a novel bacterium isolated from soil with the ability to deodorize the bad smell generated from short-chain fatty acids. Int. J. Syst. Evol. Microbiol. 54, 1997–2001. doi: 10.1099/ijs.0.03014-0
Keywords: Bacillus methanolicus, thermophile, methanol, 5-aminovalerate, alternative feedstock
Citation: Brito LF, Irla M, Nærdal I, Le SB, Delépine B, Heux S and Brautaset T (2021) Evaluation of Heterologous Biosynthetic Pathways for Methanol-Based 5-Aminovalerate Production by Thermophilic Bacillus methanolicus. Front. Bioeng. Biotechnol. 9:686319. doi: 10.3389/fbioe.2021.686319
Received: 26 March 2021; Accepted: 18 May 2021;
Published: 28 June 2021.
Edited by:
K. Madhavan Nampoothiri, National Institute for Interdisciplinary Science and Technology (CSIR), IndiaCopyright © 2021 Brito, Irla, Nærdal, Le, Delépine, Heux and Brautaset. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Trygve Brautaset, trygve.brautaset@ntnu.no
†These authors have contributed equally to this work
 Luciana Fernandes Brito
Luciana Fernandes Brito Marta Irla
Marta Irla Ingemar Nærdal
Ingemar Nærdal Simone Balzer Le
Simone Balzer Le Baudoin Delépine3
Baudoin Delépine3  Stéphanie Heux
Stéphanie Heux Trygve Brautaset
Trygve Brautaset